daniellee343
New Member
I'm working on a NUMA-related benchmark and got an issue troubling me for a week.
I use numactl to pin a metrics multiplication workload to use node0's CPU and node1's memory like this:

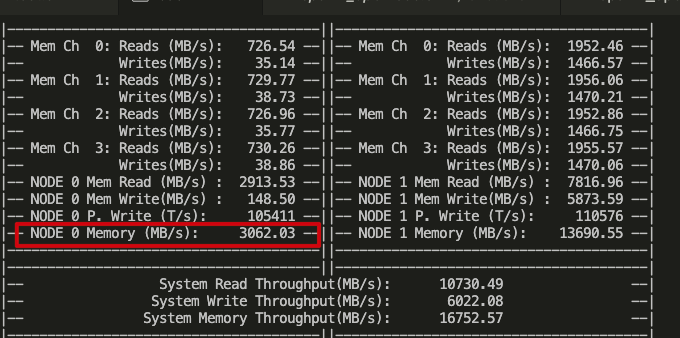
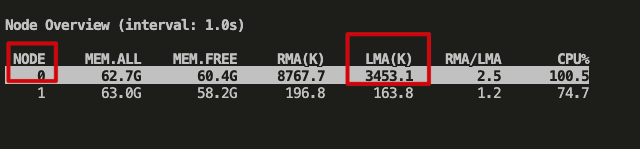
However, I observe tons of memory access still goes to node0 (throughput is high), either using pcm or numatop like this:
Then I track the pages mapping of the workload by observing /proc/<pid>/numa_maps and dump the # pages on both NUMA nodes, throughout workload running:
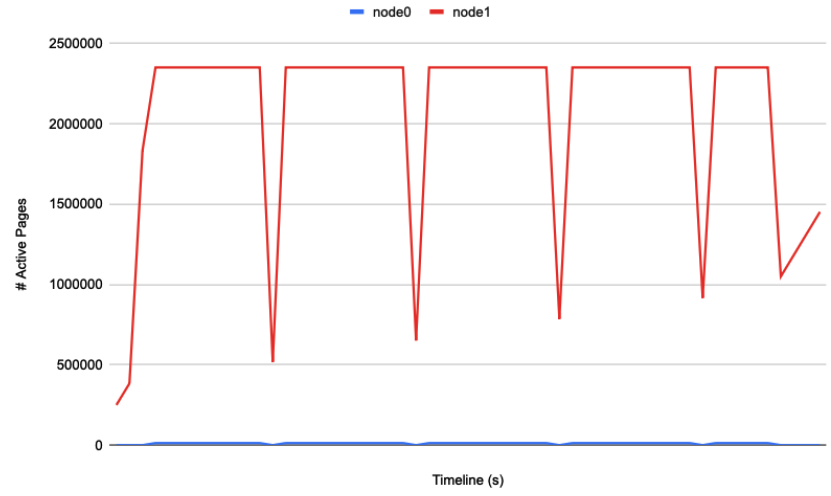
Funny thing is, node0's # active pages remains low all the way.
Then I would assume there would be dynamic libs reside on node0 during runtime, so I track those libs and evict them from memory before running. However, the result stays the same.
The question is: I see Numpy uses a well-optimized libs called OpenBLAS, but why the OpenBLAS library still has tons of small objects going to node0's memory rather than node1? On contrast, for Golang who doesn't use OpenBLAS, (almost) all its memory access goes to node1. See the figure here. Is the Linux kernel doing the trick?
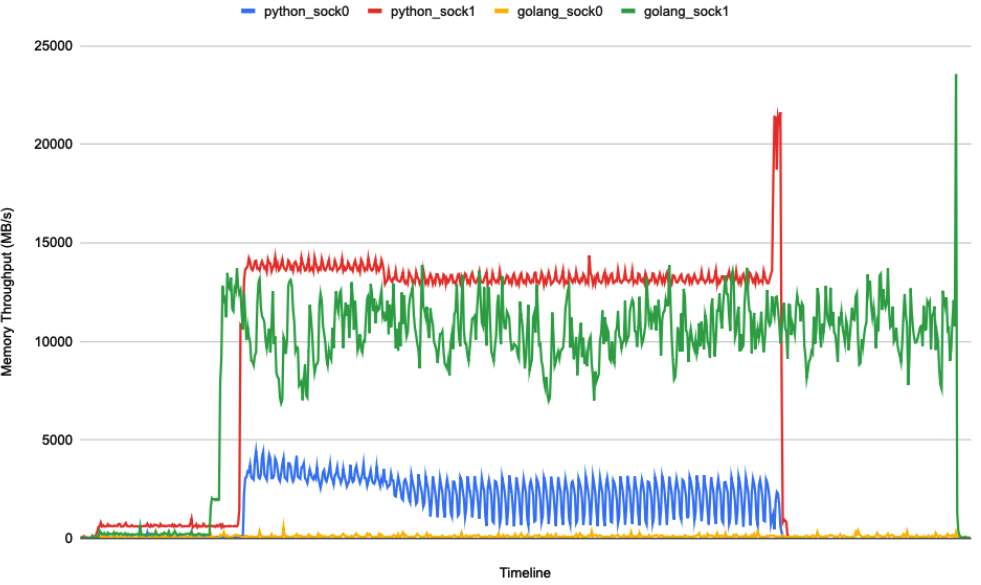
I use numactl to pin a metrics multiplication workload to use node0's CPU and node1's memory like this:
However, I observe tons of memory access still goes to node0 (throughput is high), either using pcm or numatop like this:
Then I track the pages mapping of the workload by observing /proc/<pid>/numa_maps and dump the # pages on both NUMA nodes, throughout workload running:
Funny thing is, node0's # active pages remains low all the way.
Then I would assume there would be dynamic libs reside on node0 during runtime, so I track those libs and evict them from memory before running. However, the result stays the same.
The question is: I see Numpy uses a well-optimized libs called OpenBLAS, but why the OpenBLAS library still has tons of small objects going to node0's memory rather than node1? On contrast, for Golang who doesn't use OpenBLAS, (almost) all its memory access goes to node1. See the figure here. Is the Linux kernel doing the trick?
Last edited:

